Training Neural Network Model Fitters
Mismo's neural network based fitting module uses neural networks to estimate parameter maps from MRI data with much shorter inference times than classical optimization algorithms. Unlike classical fitting, neural network estimators must be trained a-priori, typically on large amounts of simulated data (as in scheme's 1 and 2 below). A network must then be trained for each different combination of biophysical model, training scheme and data acquisition protocol, a (model, scheme, protocol) set. Once such network exists estimates can be quickly obtained for every dataset acquired with the same protocol.
Currently, three schemes of neural network based fitting are implemented: schemes 1 and 2 from ref. [1] (respectively
scheme1 and scheme2), and the self-supervised scheme introduced in ref. [2] (schemeSS).
Available Training Schemes
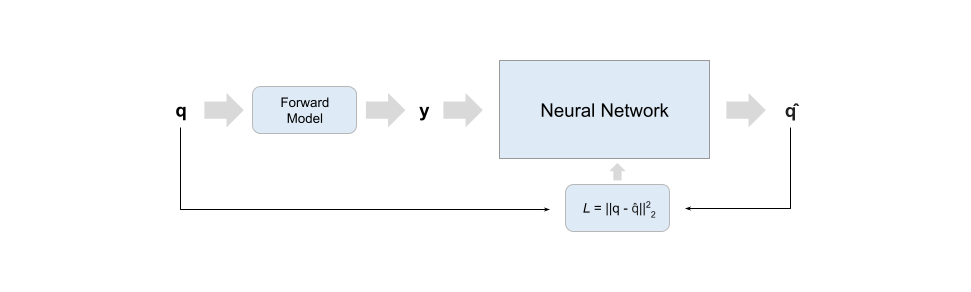
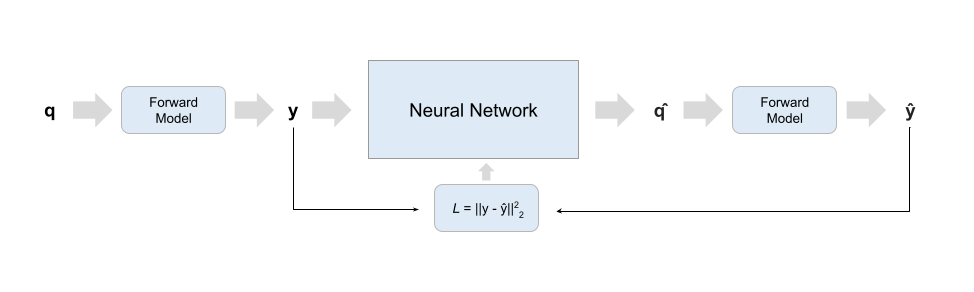
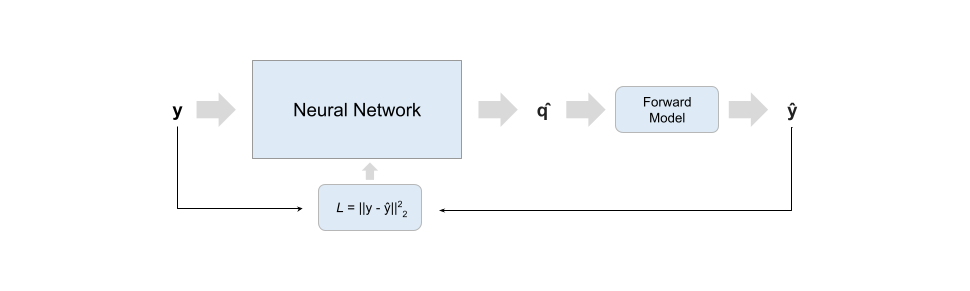
Training schemes difffer in input data and training loss. Scheme 1 and 2 networks are trained on large amounts of data generated with the specified biophysical model and can be used to fit multiple studies so long as they share the same protocol; scheme SS trains directly on acquired data and a new network is trained for each study - it thus behaves closer to a classical optimizer where the search backbone is a neural network, while schemes 1 and 2 try to approximate the forward model's inverse process.
Below are schematics for each training scheme:
Scheme 1
Scheme 2
Self-Supervised Scheme
Parameter Estimation using NNFitting
Fitters
Once a network has been trained, it can be used to perform inference on a study's data. In order to do this, a network must be initialized, input and output data must be appropriately normalized and
protocol's need to be validated. All of the necessary information for inference is thus stored alongside network weights as a clearml.Model object and logged to the Model registry. Model objects are used
to initialize fitters, instances of the ModelFitter class, which are used to perform the fitting.
Running End-to-End Parameter Estimation
Note: WIP, needs to be updated
Defining Training Parameters
The network training and optimization procedure is customisable via a set of parameter objects which the user can choose
to modify. These are NNFittingHparams, NNFittingOptions and NNHparamSearchSpace.
NNFittingHparams
The fitting hyperparameters object defines the hyperparameters for data generation and network training. In can be initialized by specifying only model_type
and training_scheme, while all other fields default to values specified via (model, scheme) specific configuration files (details below under Configuration Files).
The dataset specific parameters in_features and out_features are initialized as 0 and handled inside the training methods.
::: mismo.fitter_training.hpp_classes.NNFittingHparams options: heading_level: 4
NNFittingOptions
The fitting options object specifies system-specific options, such as where to run the training.
::: mismo.fitter_training.hpp_classes.NNFittingOptions options: heading_level: 4
NNHparamSearchSpace
The hyperparameter search space object defines which hyperparameters to optimize when running a hyperparameter optimization,
and the corresponding range of values to search. Unlike the classes above, by default all values initialize to None. Hyperparameters set
to None will not be optimized for, and all for which a range is provided will. A default set of ranges can be loaded by initializing
the object with load_defaults = True, in which case all ranges not specified by the user will be set to their default values, and all
hyperparameters for which tuning is supported will be optimized.
::: mismo.fitter_training.hpp_classes.NNHparamSearchSpace options: heading_level: 4
Configuration Files
The sections above contain references to a series of configuration files. Configuration files can be found under ./config/nnfitting, in the subdirectories nnhparams, nnoptions and nnsearchspace, respectively containing default hyperparameters, fitting options and optimization search ranges.
Default Hyperparameters
Each (model, scheme) pair requires a different set of optimal hyperparameters such as network width and depth, learning rate, weight decay etc. Since different models and training schemes may have very different optimal values for these parameters, a single set of default hyperparameters for all possible optimization scenarios would be suboptimal. On the other hand, hyperparameter optimization is too expensive to be run every time a new network needs to be trained, and since Mismo has been developed to flexibly accomodate new models and training schemes, dispatching a single set of pre-defined default values isn't feasible.
To allow for training of any (model, scheme) without defaulting to non-optimized hyperparameters, ./config/nnfitting/nnhparams is first searched for a model_scheme.yaml file containing default hyperparameter values. Then the behavior is as follows:
- If no default config exists and hyperparameter tuning is turned on: tuning will be performed for all hyperparameters regardless of provided
NNHparamSearchSpaceand the resulting optimal hyperparameters will be saved as the new defaults for subsequent runs under model_scheme.yaml - If a default config exists and hyperparameter tuning is turned on: tuning will only be performed for the specified hyperparameters (all of them when
NNHparamSearchSpaceisn't provided). - If no default config exists and hyperparameter tuning is turned off: an exception is raised prompting the user to run the training again with hyperparameter tuning enabled. In other words, a network can only be trained if a default architecture has already been established - this prevents training with suboptimal hyperparameters as a default.
- If a default config exists and hyperparameter tuning is turned off: training proceeds as normal.
- A new set of defaults can be defined by either manually editing the config file or setting
set_new_default_hpps = Truewhen hyperparameter tuning is enabled.set_new_default_hppsis automatically set toTrueif a config doesn't exist.
This ensures each (model, scheme) is trained using an optimal hyperparameter set, which is automatically saved for posterior use when optimization runs for the first time.
Default Options and Search Space
Both /nnoptions and /nnsearchspace subdirectories contain a configuration file default_config.yaml defining the default values for options and optimization ranges. The user can
set their own defaults by editing these files, or override specific default values during training, optimization or inference via keyword arguments or by instantiating
NNFittingOptions and NNHparamSearchSpace objects.
Simulating Training Data
Both scheme1 and scheme2 are trained on simulated data. A dataset is generated for a given model, acquisition protocol
and user defined SNR-level, containing n_datapoints of randomly sampled model parameters with corresponding signal sequences,
both noise-free and with added Rician noise.
from mismo.fitter_training.dmri_simulation import simulate_model_data
from mismo.models import OriginalVERDICT
model = OriginalVERDICT()
study = get_study_from_volumes(volumes)
simulate_model_data(model, study, snr=30, n_datapoints=1e6, rel_path=dataset_name)
The above code snippet samples random parameters directly from the model parameter space and computes the signal under
the defined forward model, at acquisition settings specified by the study protocol. The noisy signal is randomly
sampled from a Rician distribution where parameter $\nu$ is the groundtruth signal and $\sigma$ is calculated
using the maximum signal amplitude (typically S0) and the specified SNR.
Parameters, noise-free and noisy signal sequences are saved to the user-defined output data path under ./nn_fitting/nn_data/dataset_name.
Dataset information is stored in a dataset_info.csv file. Dataset generation hyperparameters and statistics for both noise-free
and noisy signals are also saved.
Creating Training Datasets
For Schemes 1 and 2, a SimDataset class is available which loads the simulated data. Signals are also normalized to
have zero mean and unit standard deviation using the previously computed signal statistics. Sampled model parameters are
normalized to span the [0,1] range.
from mismo.fitter_training.datasets import SimDataset
train_dataset = SimDataset(data_dir = data_path, init_ = 0, end_ = 8e5)
# Returns a training dataset starting at dataset entry 0 and ending at dataset entry 800000
train_dataset.__getitem__(0)
# Returns a tuple (torch.Tensor, Dict[str, torch.Tensor], torch.Tensor), corresponding
# to (normalized_noisy_signal, scaled_model_parameters, normalized_noise-free_signal)
For the SS Scheme, only the real data signal is available. The data class SSDataset loads all the prostate voxels from
a given study, and again normalizes their signal to zero mean and unit standard deviation.
from mismo.fitter_training.datasets import SSDataset
train_dataset = SSDataset(study, model)
# Returns a training dataset containing all prostate voxels in the study. Additionally, stores
# the model's parameter ranges for de-normalizing estimated parameters prior to the loss computation
train_dataset.__getitem__(0)
# Returns a tuple (torch.Tensor, None, torch.Tensor), corresponding to
# (normalized_signal, None, normalized_signal)
The normalization of parameters to a range serves two purposes: i) constraining the network outputs to the [0,1] range automatically scales the loss such that parameters are weighed uniformly despite large differences in order of magnitude; ii) constraining the possible parameter values is equivalent to adding a prior, which makes correct estimation easier due to the generally degenerate nature of the underlying biophysical models.
Training NN Model Fitters
New fitter neural networks can be trained for a (model, scheme, protocol) by calling get_nn_fitter. The appropriate network architecture (with the correct number of input and output nodes),
loss function and dataset class are automatically determined from the information provided.
from mismo.fitter_training.nn_train import get_nn_fitter
model_cml = get_nn_fitter(model_name, study, training_scheme, ***options)
This method first searches the model registry for an existing published Model trained for a matching (model, protocol, scheme), and returns a clearml.Model object which can be used to instantiate a ModelFitter. If no published Model for (model, protocol, scheme) can be found, a new neural network is trained and the corresponding Model object if logged to the registry. The behavior of this function can be modified in a series of ways:
- If a training dataset already exists, the user can provide its relative path to skip the data generation process - otherwise data is generated with default SNR and number of datapoints, configurable by providing the key word
arguments
n_datapointsandsnr. This input has no effect when training withschemeSS - Training a new model regardless of whether a matching published one already exists can be done by setting
overwrite = True. - Providing
nn_hparamsand/ornn_optionsallows for user-defined hyperparameters to be used in the training process. When none are provided, default values are loaded from a config file if one exists. (for more details see Defining Training Parameters) - setting
tune_hyperparameters = Truewill perform a hyperparameter search prior to training the network. ANNHparamSeachSpaceobject can be provided specifying which hyperparameters to optimze and corresponding ranges; otherwise all hyperparameters will be optimized, with default search ranges as defined via ./config/nnfitting/nnsearchspace/default_config.yaml - when running hyperparameter optimization, setting
set_new_default_hpps = Truewill save the final hyperparameters to the (model, protocol, scheme) default configuration file (more details can be found below under Configuration Files).
💡 Publishing Models: by default, only published Models are queried, and a Model must be published manually by the user. Models should only be published when they are the current optimal version for the respetive (model, scheme, protocol). If you wish to set a new Model as the default for (model, scheme, protocol), but a published one already exists, you must delete the existing published model and publish the new one instead. This ensures only one Model per (model, scheme, protocol) exists for common use.
Performing Model Inference
Once a model has been trained, it can be used to compute estimated parameter maps for any study acquired with the protocol
the fitter has been trained for. This is done via the ModelFitter class. All that is needed to initialize a ModelFitter object is a clearml.Model object, which contains all network and data information.
from mismo.fitter_training.nn_fitters import ModelFitter
from mismo.fitter_training.utils import find_fitter
fitter_clm = find_fitter(protocol, hparams)
fitter = ModelFitter(fitter_clm) #Optionally: ModelFitter(fitter_clm, device = "gpu")
parameter_maps = fitter.fit_volumes(study)
# Returns a dictionary of study.data.shape volumes containing the estimated parameters for each prostate voxel
💡 Finding Models in the Registry: the ModelFitter class was not designed to be used directly but instead within
fit_nn_optimizeras detailed in section End-to-End Parameter Estimation. However, if one wants to instantiate a ModelFitter object, the following methods can be used to find a Model in the registry: callingget_nn_fitter(see Training NN Model Fitters), which will train a fitter if a published one doesn't already exist; or usingfind_fitter, which simply queries the registry, and can be used to fetch non-published models by settingonly_published = False
References
[1] Deep learning model fitting for diffusion-relaxometry: a comparative study .Francesco Grussu, Marco Battiston, Marco Palombo, Torben Schneider, Claudia A. M. Gandini Wheeler-Kingshott, Daniel C. Alexander
[2] Sen, Snighda; Singh, Saurabh; Pye, Hayley; Moore, Caroline; Whitaker, Hayley; Punwani, Shonit; Atkinson, David; Panagiotaki, Eleftheria; Slator, Paddy; (2023) Self-Supervised Model Fitting Of VERDICT MRI In The Prostate. In: Proceedings of the 2023 ISMRM & ISMRT Annual Meeting & Exhibition.