Concepts & Datastructures
Mismo includes implementations of state-of-the-art microstructure models as well as multiple different methods to fit MRI data to microstructural models.
Datastructure: Studies
Mismo depends on the datastructure Volume from the vicom package.
Mismo however defines another datastructure called Study.
A Study is a collection of Volume objects belonging to a single patient
acquisition setting.
Three lists of volumes make up a study:
- DWI Volumes:
study.dwi - T2 Volumes:
study.t2 - Mask Volumes:
study.mask
We separate the volumes into these three partitions for easier access and less
boilerplate code. For example, the microstructure model fitting process only
accesses the dwi and mask partitions. The segmentation however only
accesses t2, whereas the registration process only accesses all three
partitions.
The Study moreover validates provided volumes as well as provides a few
helpful functions commonly used in the processing steps.
Datastructure: Modeling Framework
Mismo attempts to define a framework for microstructure modeling. Traditional microstructure models are typically defined by an equation expressing the expected signal as a function of the microstructure parameters. An overview over a number of microstructure models can be found in 1.
We implement a modeling framework which can be visualized as follows (ignoring any UML correctness):
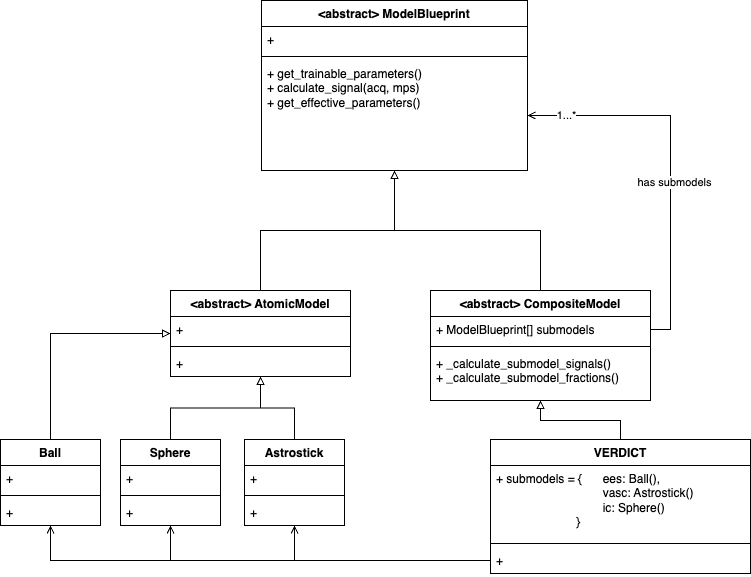
The highest-level class is the ModelBlueprint, which defines the three core
functions every model implementation has to provide.
References
::: mismo.data.data_models options: heading_level: 3 members:
- Study
- get_study_from_volumes
::: mismo.models.ModelBlueprint options: heading_level: 3
Footnotes
-
Panagiotaki, Eleftheria, et al. "Compartment models of the diffusion MR signal in brain white matter: a taxonomy and comparison." Neuroimage 59.3 (2012): 2241-2254. ↩